Portworx & Red Hat Hands-on Labs Register Now
Generative AI has been top of mind for individuals and organizations for more than a year now. It has certainly brought the whole discipline of artificial intelligence and machine learning to the forefront and organizations are now hiring and enabling data scientists and AI/ML engineers to create better user experiences for their customers. This can include everything from using GPT-based models to create custom chat bots to building machine learning models that can be deployed inside manufacturing facilities to monitor the entire supply chain process. It is at this crossroads that organizations have to figure out what the right tools for the job are, and how they can equip these highly skilled individuals to do what they should be doing instead of drowning in the minutiae of infrastructure management.
In this blog, we will talk about how Kubernetes and Portworx by Pure Storage can help data scientists accelerate their training and experimentation cycles to build ML models faster. As we know, machine learning is the process of using data to answer questions. You use data to train ML models, and once it is trained, these models can be leveraged to predict or infer answers for any future questions. Let’s look at a simple example from the real world, companies like Annalise AI are building models to examine chest x-rays, to help accelerate the diagnosis of patients in areas that don’t have radiologists available to help everyone.
Jupyter Notebooks
Choosing the right tool for the job is absolutely critical. Ask anyone who has tried to open a can using a table knife. Jupyter notebook is the preferred tool of choice – it’s like an IDE that allows data scientists or data analysts to perform tasks like data cleansing and transformation, data visualization, data enrichment, exploratory data analysis, machine learning, and deep learning.
Jupyter notebooks have a front end browser-like interface that allow data scientists to use different python, R, etc SDKs to manipulate data and train their models, and a back-end kernel that has different packages installed which runs the code and returns results back to the web page. This allows for rapid experimentation and prototyping, as they don’t have to save different versions of the source code and find an environment to run it when changes are needed. Jupyter notebooks provide the perfect sandbox needed by data scientists, and are widely adopted in organizations like Apple, Google, LA Times, Clemson University, etc.
Although Jupyter notebooks are simple to use and easy to install on any machine that has python installed including mac or linux workstations, data scientists won’t be as productive due to resource limitations on their laptops. Local Jupyter notebooks restrict the sharing of research between teams and users, and make the overall process disjointed and inefficient.
Kubernetes to the rescue
Now that we know the advantages of using Jupyter notebooks and the limitations of running them locally, let’s see how we can scale these notebooks by using Kubernetes. Kubernetes has become the de-facto orchestration platform for a majority of new applications, and because of its extensibility, it looks like a promising platform to run machine learning workloads as well. There are already public references around how companies like OpenAI and Anthropic are using Kubernetes to build and train their Large Language models, while at the same time vendors like NVIDIA are supporting the use of containers and Kubernetes using services like the NVIDIA NGC catalog. So, let’s look at some of the benefits Kubernetes provides that enable organizations to run efficient ML pipelines.
- Shared Infrastructure Resources: Instead of dedicating individual server resources to data scientists, Kubernetes allows platform teams to increase their resource utilization by using a shared platform for multiple data scientists. Platform teams can spin up individual Kubernetes clusters for each team of data scientists or deploy and manage a multi-tenant Kubernetes cluster for multiple teams. With coveted resources like GPUs, a shared platform like Kubernetes can help organizations achieve higher resource utilization. Based on the GPU that organizations have access to, they can leverage multipie GPU concurrency choices, from ‘time slicing’ – which allows dividing GPU access into small time intervals, to ‘Multi-Instance GPUs (MIGs)’ – which allow a single GPU to be partitioned into multiple instances, to ‘Virtual GPUs (vGPU)’ – which allows users to share the power of the same physical GPU across different virtual machines or Kubernetes worker nodes.
- On-demand Access: Kubernetes allows users to deploy their Jupyter notebooks when they need them and it then orchestrates these notebooks as pods, ensuring they have access to the compute, memory, and GPU resources requested. Because of extensions like the Cluster Autoscaler and the open source Karpenter project, Kubernetes can spin up additional worker nodes to support the incoming requests from data scientists. This also allows organizations to control costs, as they don’t need to have previously overprovisioned Kubernetes clusters sitting idle, but can scale up their clusters when needed.
- Portability: Kubernetes has become the platform of choice when it comes to avoiding vendor lock-in. All major cloud providers and independent software vendors have a flavor of CNCF conformant Kubernetes distribution that they can provide to their customers to run their containerized applications. This means that an organization can choose the cloud provider they want, without having to rearchitect or change the way they build and operate their machine learning workflows because of the consistent orchestration layer offered by Kubernetes.
- Repeatability: As part of each Jupyter notebook, there are different kernel versions, package, and SDK versions that need to be configured for the code to be executed. Because of the benefits offered by containers and Kubernetes, this allows users to package all of the dependencies in a specific container image, and then deploy it consistently across any Kubernetes cluster. This way, data scientists don’t have to worry about managing kernel versions across different environments. Individual notebook container images can be stored in either a public or a private container registry and pulled into any Kubernetes cluster.
Although Kubernetes offers all of these benefits, there is still a learning curve associated with it. Organizations shouldn’t expect their data scientists to learn more about Kubernetes and become experts. They are being paid to build highly accurate models and not worry about authoring yaml files and figuring out how to configure a statefulset object in Kubernetes. This is where platform teams inside organizations should take on the onus of extending the platform they have built for their developers and also support data scientist teams. In the next section, we will look at one such project in the open source community called Kubeflow which provides a self-service way for data scientists to spin up Jupyter notebooks and build ML pipelines.
Kubeflow
Kubeflow is a Cloud Native Computing Foundation (CNCF) incubating project that aims to be the machine learning toolkit for Kubernetes. It allows data scientists to run ML workflows on Kubernetes easily, and in a scalable fashion. Kubeflow has different components that can be used during different stages of the ML pipeline, but in this blog we will specifically focus on its ability to dynamically provision notebooks and pipelines.
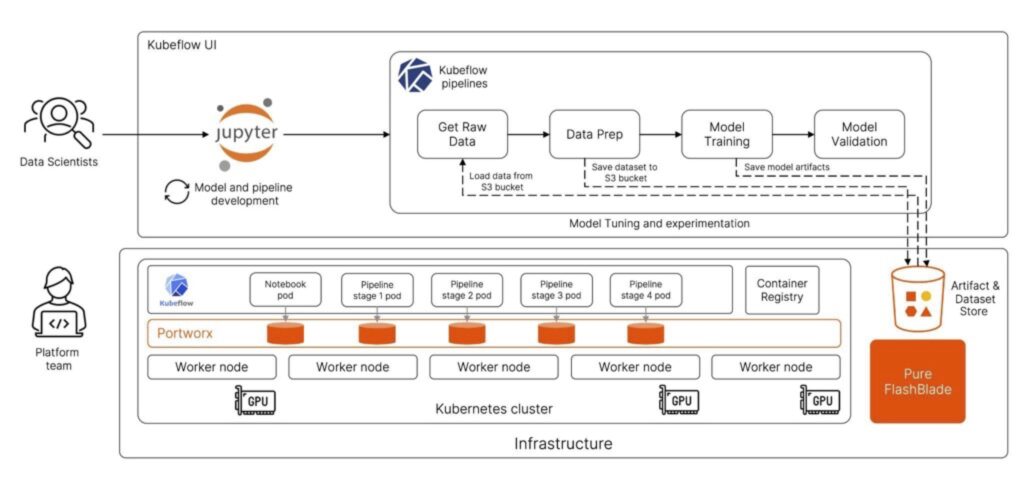
Kubeflow notebooks allow users to simply navigate to the UI, and provision dedicated development environments that can be based on either Jupyter, RStudio, or Visual Studio Code. During the notebook creation process, the user can select the specific container image to use which will have pre-packaged binaries needed, along with a request for the amount of CPU, Memory, Storage, and GPU resources needed, and hit launch!
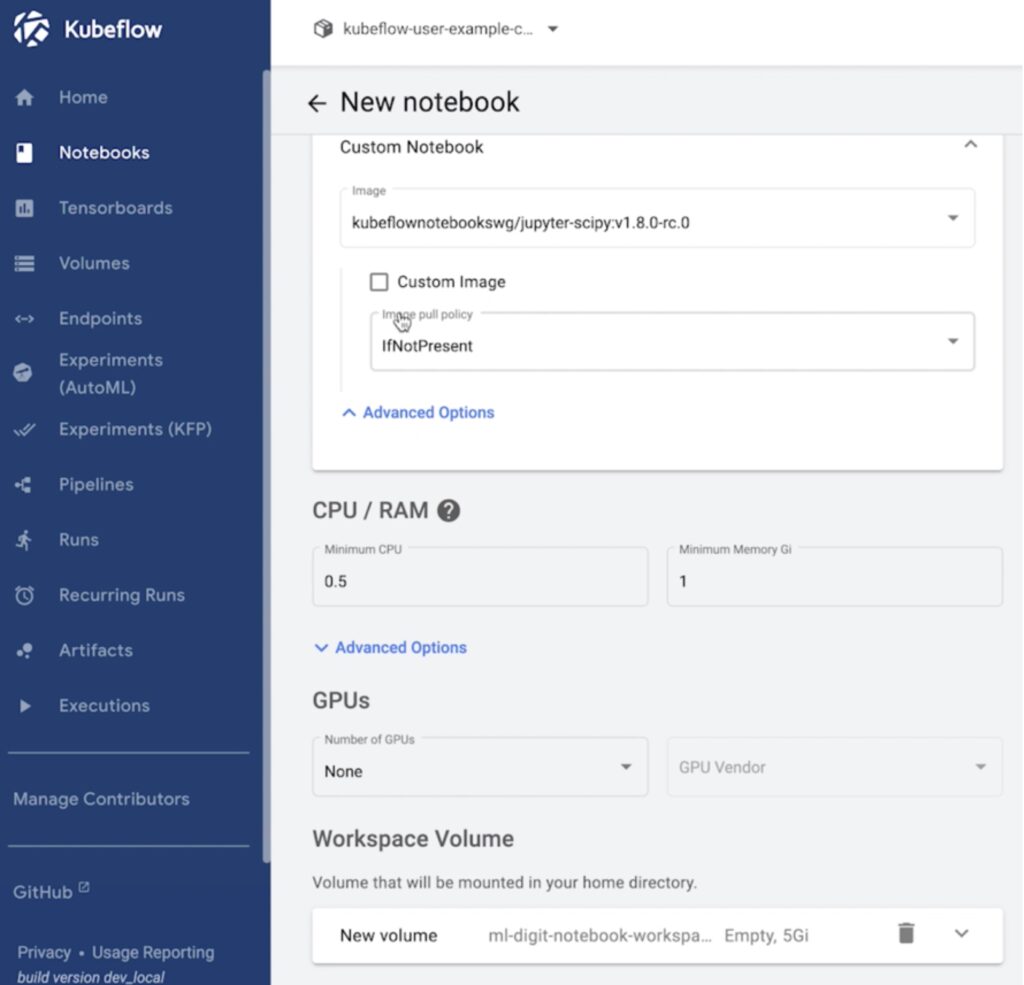
At this point, Kubeflow deploys a pod on the backend Kubernetes cluster and gives the user access to the Jupyter notebook.
So, all the data scientist needed to do was go to a GUI and issue a request for a Jupyter notebook without having to worry about deploying it locally or figuring out the resources needed to run the notebook. Once Kubeflow completes the notebook deployment, the data scientists can start using the hosted notebook IDE to build and train their models.
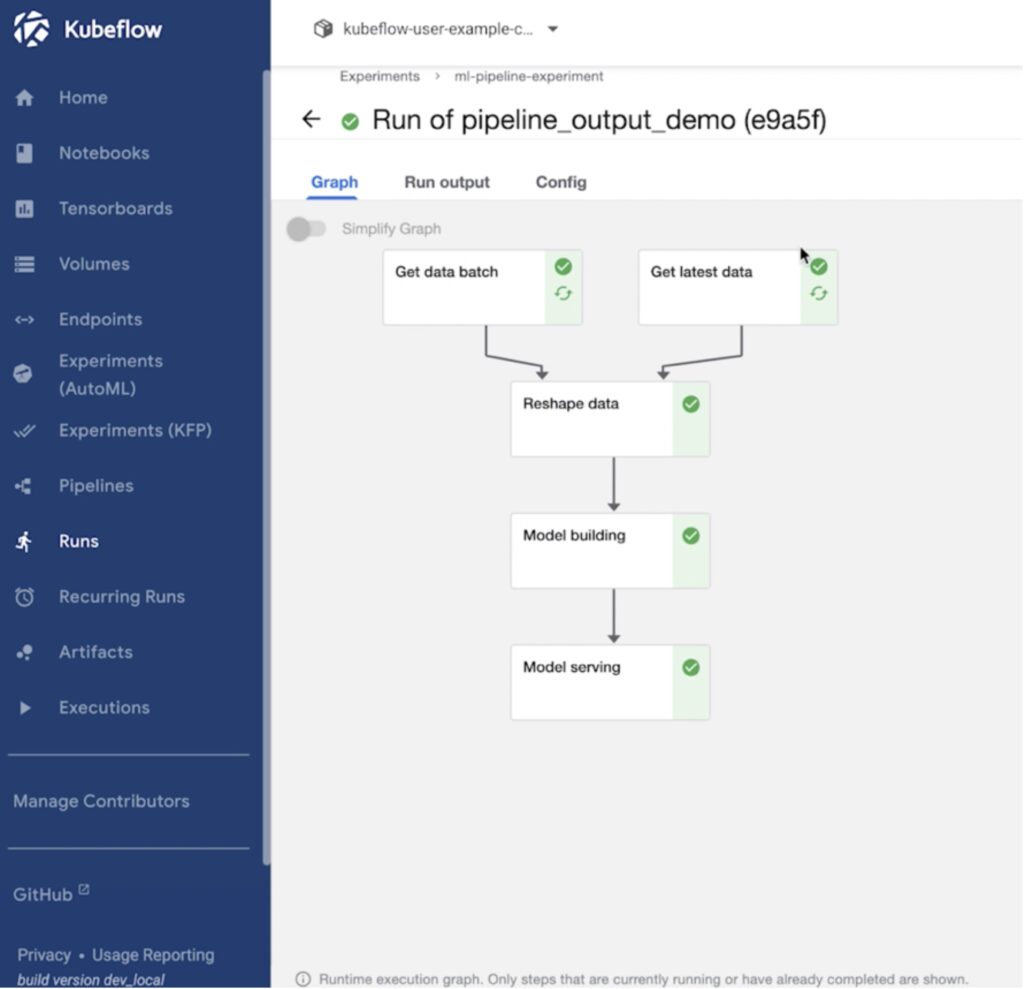
Another useful feature in Kubeflow is the ability to create pipelines and run experiments to fine tune ML models. Data scientists can use their Jupyter notebooks to create a pipeline object for their Directed Acyclic Graphs (DAG), or they can generate a yaml file for the pipeline object from the Jupyter notebook and use that to create a new Kubeflow pipeline object.
Once a pipeline is created, users can run individual experiments and perform hyperparameter tuning, or they can also share their pipelines with other users on the team and everyone can run their own experiments using Kubeflow. During each experiment run, Kubeflow spins up the needed containers and pods on the backend Kubernetes cluster with the amount of CPU, memory, GPU, and storage resources needed to run each stage of the DAG.
Portworx and Pure Storage
Kubeflow and Kubernetes definitely make things easier for data scientists and platform engineers, but they alone won’t help organizations build a better platform. In this section, we will cover how Portworx and Pure Storage augment the capabilities offered by Kubeflow and Kubernetes.
Data is the most critical part of training any model. Some might say it is GPU capacity, but if bad data is used it will result in inaccurate models being trained, which defeats the purpose of using machine learning to solve business problems.
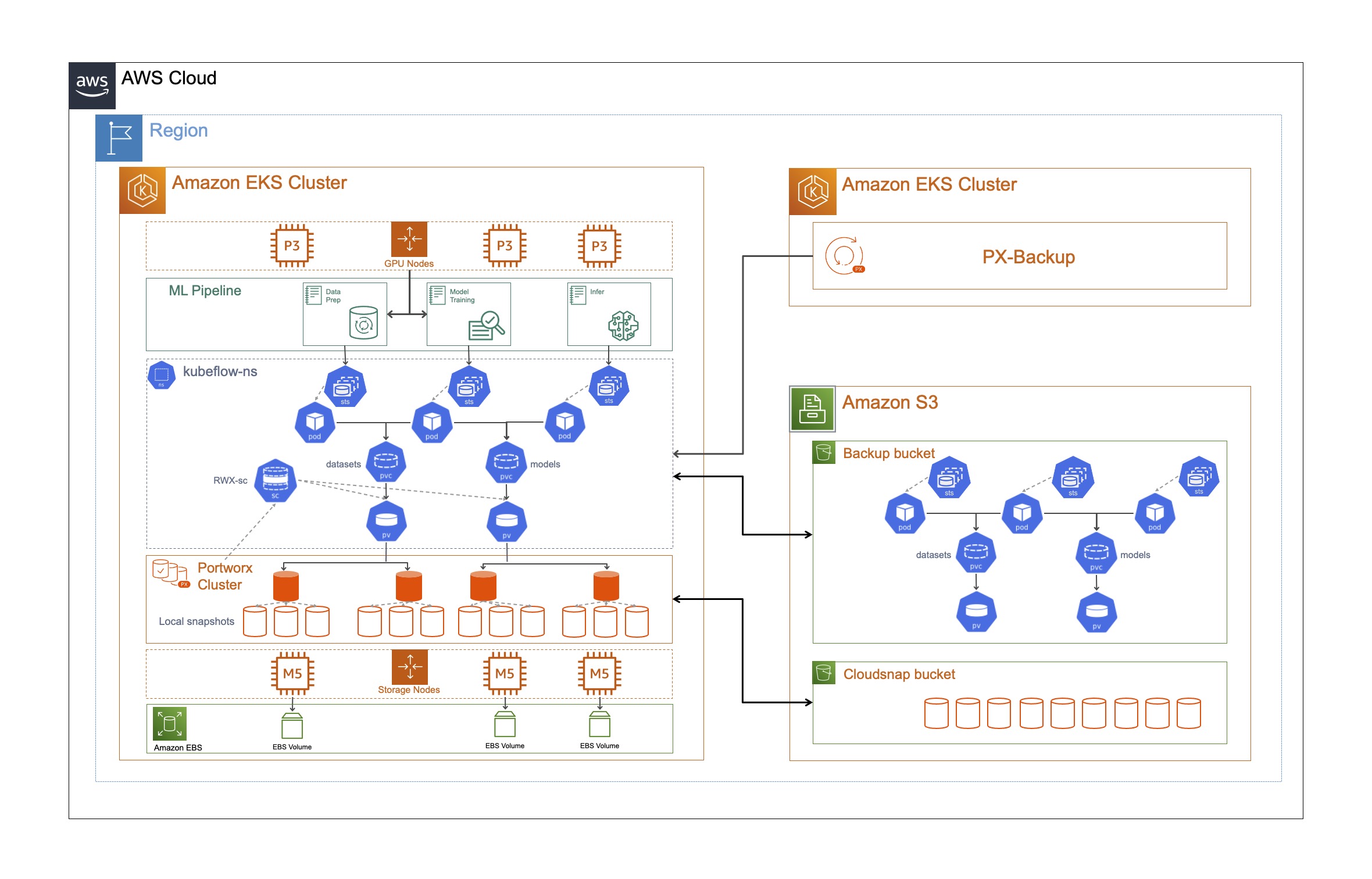
Scale-out Object Storage: When training custom models, Pure Storage FlashBlade provides a scale-out storage platform that efficiently powers modern unstructured data needs without complexity. Pure Storage FlashBlade offers a fully S3-compatible on-premises platform that organizations can use to store their raw data.
Dynamic Provisioning of Block and File Volumes: Portworx supports the dynamic provisioning of block and file based volumes for notebooks created through the Kubeflow UI. This allows data scientists to provision storage needed for their notebooks on a self-service basis instead of having to request for storage from their platform team. File volumes can be used by multiple notebooks at the same time, so data scientists can either treat them as home directories across multiple notebooks of their own, or file volumes can also be shared across notebooks from different data scientists, allowing them to share access to trained data or a specific dataset, rather than having to download TBs of data every time they want to manipulate or use that dataset.
High Availability and Replication: Failures should always be anticipated, especially during long running jobs. Portworx allows platform engineers to set replication factors at the Kubernetes layer, so all your data is replicated across different nodes in the cluster, which avoids any data loss in case of node failures or rack failures during model training.
Local Snapshots and Cloudsnaps: Portworx allows platform engineers to create snapshot policies that automatically take snapshots of the persistent data and store it in any S3-compatible storage array like Pure Storage FlashBlade. But in addition to this, Portworx also offers its own Python SDK, that can be used by data scientists as part of their notebooks, to take snapshots and save the state of their notebook once they reach a specific milestone.
Multi-tenant Clusters: As we discussed earlier in the blog, resource utilization is a big challenge when it comes to compute resources for AI/ML. With Portworx, platform engineering teams can create different classes of service when it comes to storage services and allow data scientists and other personas within an organization to share the same underlying infrastructure, without anyone starving for resources.
Conclusion
Reinventing the wheel is usually not a great idea. Instead of building dedicated infrastructure silos for data scientists inside an organization, think about how existing platforms can be extended to benefit from economies of scale inside an organization. Projects like Kubeflow on Kubernetes allow platform engineering teams to serve data scientists using the same infrastructure stack and automation built for developers building containerized applications.
We also recorded a quick demo around how Portworx and Pure Storage can help organizations run their Jupyter notebooks at scale.
Portworx offers a free trial for users who want to get their hands dirty and deploy Portworx on their own Kubernetes cluster. Just create a new account on Portworx Central, and start using Portworx today!
Share
Subscribe for Updates
About Us
Portworx is the leader in cloud native storage for containers.
Thanks for subscribing!

Bhavin Shah
Sr. Technical Marketing ManagerExplore Related Content:
- jupyter
- jupyterhub
- kubeflow
- machine learning